A survey of non-redundant protein domains shows that natural protein domains are strongly cross linked by minimally frustrated contact networks comprising about 40% of the total contacts [11]. Only a minority (about 10%) of the native interactions are found to be 'highly frustrated', and they typically cluster at the protein surface. The remaining 50% are 'neutral' and are ramdomly distributed in the structure (not draw).
The highly frustrated interactions that, in principle, might conflict with the robust folding of the domain, seem to reflect evolutionary constraints other than folding and often correspond to physiologically relevant sites.
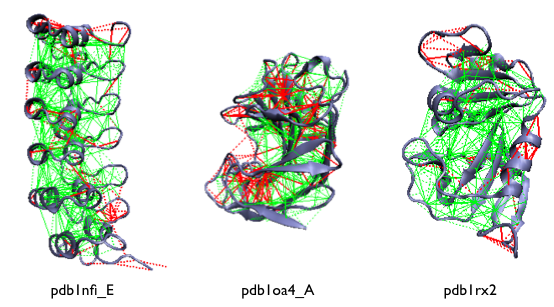
Examples of the localized frustration and minimally frustrated networks in protein structures. The protein backbone is displayed as blue ribbons, the direct inter-residue interactions with solid lines and the water-mediated interactions with dashed lines. Minimally frustrated interactions are shown in green, highly frustrated contacts in red, neutral contacts are not drawn.
A statistical survey shows that these sites do co-localize with regions involved in the formation of heterodimeric protein assemblies [11].
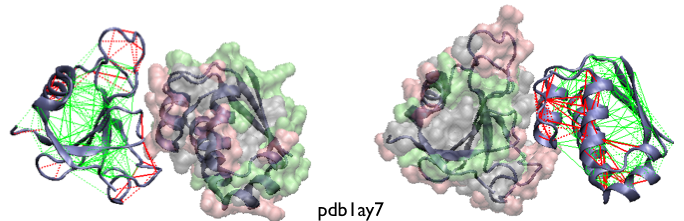
Example of localized frustration patterns in protein assemblies. The interactions in one monomeric partners are colored according to the contact configurational frustration index while the other partner's surface is colored according to the single-residue level frustration index. The frustration indices are shown as calculated for the unbound monomers. Complementary views of the same complex is shown.
Another survey shows that highly frustrated sites do co-localize with regions involved in allosteric transitions [16].
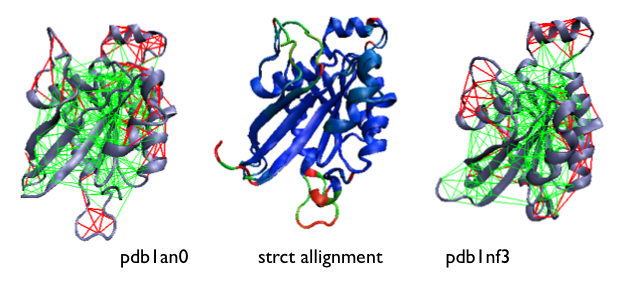
Example of the localized frustration and minimally frustrated networks in allosteric proteins [16]. A structural allignment of both experimentally determined conformations is shown at the center, coloured according to their deviation (blue low, red high). The configurational frustration pattern of the individual conformations are shown at the sides.
Every protein has its own history (and story) to tell. The possibility of localizing and quantifying the energetic frustration present in protein molecules allows one to probe lower hierarchies of the energy landscape, manifested as the exploration of the configurational substates defined by the local roughness. Molding of this roughness can have profound effects on the structural transitions and is thus likely to have functional consequences[9, 10]. Particular examples can be very interesting, but results should be taken carefully. Performing a statisticall survey (of homologs, mutants, etc) is encouraged.
Enzyme active sites are often found frustrated. Several Beta-lactamases are shown.
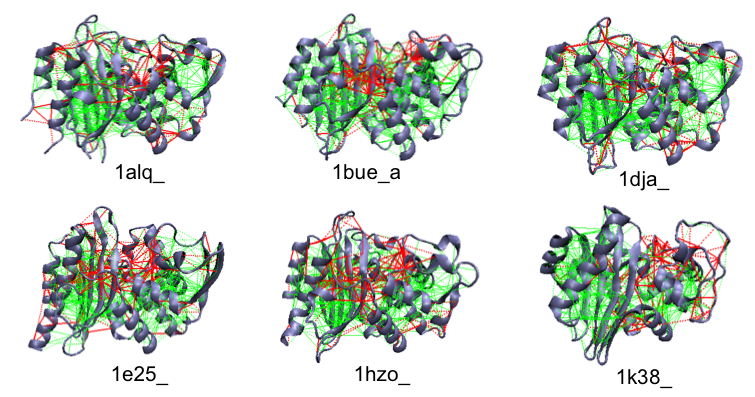
Example of the localized frustration and minimally frustrated networks in beta-lactamase enzymes. The active site (roughly at the center) is shown to have highyl frustrated interactions.
Highyl frustrated regions found in the native state contribute to the stabilization of folding intermediates. [15, 14]
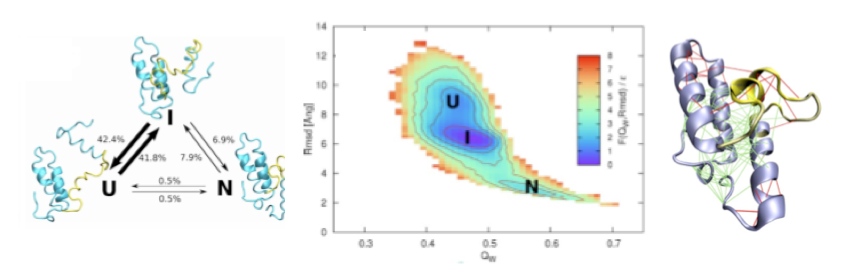
A non-native folding intermediate is formed by the Im7 protein. Analysis of structures derived from folding simulations suggest that the intermediate ensemble (on the right) is stabilized by non-native minimally frustrated contacts, while the native ensemble is locally destabilized by a highly frustrated interactions (on the left).
D34: a 12-ankyrin repeat fragment of ankyrinR [15]
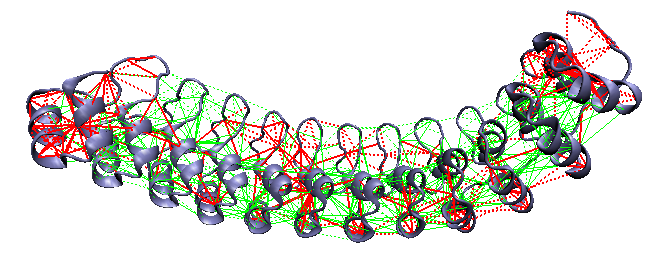
Local frustration on D34: a 12-ankyrin repeat fragment of AnkyrinR. This protein folds through an intermediate with roughly half of the repeats folded. The highly frustrated regions are located at the ends, and in a central region between repeats 5 and 6, in an otherwise strongly crosslinked minimally frustrated network. These breaks occur where the experiments suggest the intermediate's folding boundary is.